Plant formins homepage
Note: details on alternative gene predictions from Uniprot will be added later.
AtFH15a |
Phylogenetic class II |
Structure type E |
|
AtFH15a |
Phylogenetic class II |
Structure type E |
|
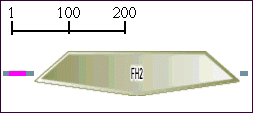
Key to domain abbreviations can be found here.
>AtFH15a
MSLVEISGSDAMAAPMPGRVPPPPPRPPPMPRRLPPMFDAFDHTGAGMVW
GFPRPAKKRASLKPLHWVKITSDLQGSLWDELQRRHGDSQTAIELDISEL
ETLFFVEAKPEKIRLHDLRRASYRVFNVRSYYMRANNKVINLSMPLPDMM
TAVLAMDESVVDVDQIEKLIKFCPTNEEMELLKTYTGDKAALGKYEQYLL
ELMKVPRLEAKLRVFSFKTQFGTKITELKERLNVVTSACEENLLLIHQVR
SSEKLKEIMKKIPCLGNTSNQGPDRGKTFLSPVEFKLDRLSVKRMHYFCK
LKEIMKKIPCLGNTSKSNPRVGVKLDSSVSDTHTVKSMHYYCKVLASEAS
ELLDVYKDLQSLESASKIQVKSLAQNIQAIIKRLEKLKQELTASETDGPA
SEVFCNVCWFFVRLMI
AtFH15b |
Phylogenetic class II |
Structure type C |
|
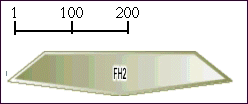
Key to domain abbreviations can be found here.
>AtFH15b (AtF20)
MTLFNFIKLFKKAHEENVKQADLEKKKAMKQIDLRRANDTEIMLTKVNIP
LADMMAAVLGMDEYVLDVDQIENLIRFCPTKEEMELLKNYTGDKATLGKC
EQLAKAKAPLKEHFRVINAFPSLTPQYFLEVMKVPGVESKLRAFSFKIQF
GTQIAELNKGLNAVNSACEEVRTSEKLKEIMANILCMGNILNQGTAEGSA
VGFKLKSLLILSDTCAPNSKMTLMHYLCKVLASKASDLLDFHKDLESLES
ASKIQLKSLAEEIQAITKGLEKLNKQLTASESDGPVSQVFRKVLKDFISM
AETQVATVSSLYSSVVMFVNLLLLVDSKCLTFYNVFDLEITTTLLSFIRL
FKKAHEENVKQADLDKNKDAKEAEMEKTK
AtFH15 |
Phylogenetic class II |
Structure type E |
|

Key to domain abbreviations can be found here.
>AtFH15 - original prediction from genome annotation GenBank NP_196382.2
MSLVEISGSDAMAAPMPGRVPPPPPRPPPMPRRLPPMFDAFDHTGAGMVWGFPRPAKKRASLKPLHWVKI
TSDLQGSLWDELQRRHGDSQTAIELDISELETLFFVEAKPEKIRLHDLRRASYRVFNVRSYYMRANNKVI
NLSMPLPDMMTAVLAMDESVVDVDQIEKLIKFCPTNEEMELLKTYTGDKAALGKYEQYLLELMKVPRLEA
KLRVFSFKTQFGTKITELKERLNVVTSACEEVRSSEKLKEIMKKIPCLGNTSNQGPDRGKSSVVDKNLSF
SSGIQLKEIMKKIPCLGNTSKSNPRVGVKLDSSVSDTHTVKSMHYYCKVLASEASELLDVYKDLQSLESA
SKIQVKSLAQNIQAIIKRLEKLKQELTASETDGPASEVFCNTLKDFISIAETEMATVLSLYSVVRKKADA
LPPYFGEDPNQCPFEQLTMTLFNFIKLFKKAHEENVKQADLEKKKAMKQIDLRRANDTEIMLTKVNIPLA
DMMAAVLGMDEYVLDVDQIENLIRFCPTKEEMELLKNYTGDKATLGKCEQLAKAKAPLKEHFRVINAFPS
LTPQYFLEVMKVPGVESKLRAFSFKIQFGTQIAELNKGLNAVNSACEEVRTSEKLKEIMANILCMGNILN
QGTAEGSAVGFKLKSLLILSDTCAPNSKMTLMHYLCKVLASKASDLLDFHKDLESLESASKIQLKSLAEE
IQAITKGLEKLNKQLTASESDGPVSQVFRKVLKDFISMAETQVATVSSLYSSVGKNADALAHYFGEDPNH
YPFEKVTTTLLSFIRLFKKAHEENVKQADLDKNKDAKEAEMEKTK