To verify or correct ORF prediction, available EST or cDNA
sequences have been aligned to the genomic locus (text
file of the alignment here):

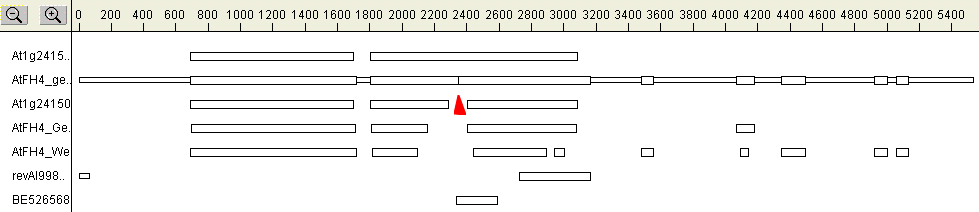
The original ORF prediction misses some conserved sequences, and the 2nd intron at the position of the conserved G-N-X-M-N motif (red line) is poorly supported by other prediction programs (WebGene, GenScan). Moreover, cDNA sequence suggests that this intron is not spliced out of the transcript. Although the unspliced "intron" contains an in-frame stop codon, an extra G (red arrow) is present in the cDNA sequence. Since insertion of this extra base restores the reading frame within a highly conserved portion of the protein, an error in the genomic sequence is highly likely (see Cvrckova et al. 2004 for more detail).
The extra exons predicted by Genscan and Webgene) were ignored, since they are not supported by cDNA.
A predicted cDNA sequence has been assembled, keeping the 2nd intron unspliced
and introducing the extra base (download here).